Use of gene expression profiles for prediction of biological activity of compounds
I mainly work on the development of an artificial intelligence model aimed at predicting the effects of chemical compounds on cancer cell lines. The goal is to virtually identify molecules that could serve as promising candidates for the development of new drugs. In this context, I collaborate closely with Prof. Quentin Fournier, Associate Researcher at Mila and Adjunct Professor at DIRO, as well as with his PhD student, Lola Le Breton.
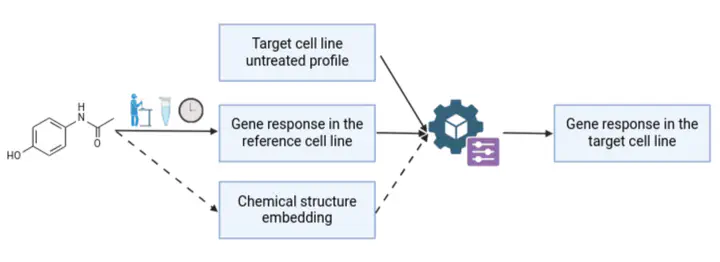
We define a “treatment” as the exposure of a cell (or a cell line) to a given molecule, at a specific dose and for a defined duration. Our hypothesis is that the gene expression changes induced by a treatment provide an informative representation of that treatment. We refer to the collection of these changes as a delta profile, defined as the difference between the treated and untreated gene expression profiles.
We train a model to predict the delta profile of a target cell line subjected to a treatment using two inputs: (i) the untreated gene expression profile of the target cell line, and (ii) the delta profile observed in a reference cell line exposed to the same treatment, but unrelated to the target cell line. This approach, based on a direct biological representation of the treatment, achieves better performance than methods relying solely on the chemical structure of the molecule.
We nevertheless investigate the potential contribution of chemical structure embeddings derived from foundation models, in order to assess whether they can further improve model performance.